求助主题PMID 33181087
需求说明全文
求助时间2021-06-29 22:40
Transcriptional Regulation and Characterization of the Promoter Region of the Human ABCC6 Gene
Qiujie Jiang1,3, Yasushi Matsuzaki1,3, Kehua Li1and Jouni Uitto1,2
ABCC6, a member of the adenosine 5'-triphosphate-binding cassette family of genes, encodes multidrugresistance-associated protein 6, a putative transmembrane transporter expressed primarily in the liver and to asignificantly lower extent in other tissues. Mutations in ABCC6 result in pseudoxanthoma elasticum, a multi-system heritable connective tissue disorder with variable phenotypic expression. To examine the transcriptionalregulation and tissue-specific expression of this gene, we cloned 2.6 kb of human ABCC6 promoter anddeveloped a series of 5'-deletion constructs linked to luciferase reporter gene. Transient transfections in anumber of cultured cell lines of perse origin identified a specific NF-kB-like sequence ( 235/ 226), whichconferred high level of expression in HepG2 hepatoma cells, inferring liver specificity. The functionality of thepromoter fragments was confirmed in vivo by tail vein injection followed by luciferase reporter assay. Testing ofselected cytokines revealed that transforming growth factor (TGF)-b upregulated, while tumor necrosis factor(TNF)-a and interferon (IFN)-g downregulated the promoter activity in HepG2 cells. The responsiveness to TGF-bwas shown to reside primarily within an Sp1/Sp3 cognate-binding site at 58 to 49. The expression of theABCC6 promoter was also shown to be markedly enhanced by Sp1 protein, as demonstrated by cotransfectionof ABCC6 promoter–luciferase constructs and an Sp1 expression vector in Drosophila SL2 cells, which aredevoid of endogenous Sp1. Furthermore, four additional transcription factors, with their cognate-bindingsequences present in DNA, were shown to bind the 2.6-kb promoter fragment by protein/DNA array.
Collectively, the results indicate that human ABCC6 displays tissue-specific gene expression, which can bemodulated by proinflammatory cytokines. These findings may have implications for phenotypic expression ofheritable and acquired diseases involving abnormality in the ABCC6 gene.
Journal of Investigative Dermatology (2006) 126, 325–335. doi:10.1038/sj.jid.5700065; published online 22 December 2005
INTRODUCTION
ABCC6 (GenBank nos. U91318 and AF076622) encodesMRP6, a member of the multidrug resistance-associatedprotein (MRP) family (Borst et al., 1999). The interest inABCC6/MRP6 has been recently heightened by demonstra-tion of mutations in this gene/protein system in families withpseudoxanthoma elasticum (PXE), a multi-system disorderaffecting the elastic structures in the skin, the eyes, and thecardiovascular system (Le Saux et al., 2001; Ringpfeil et al.,2001a; Uitto et al., 2001). The clinical manifestationsinclude loose and sagging skin, development of angioidstreaks in the eyes, which can lead to loss of visualacuity, and development of early cardiovascular disease(Ringpfeil et al., 2001a; Uitto et al., 2001). As the primary siteof ABCC6 gene expression is the liver, it has been suggestedthat PXE is primarily a metabolic disorder due to alteredfunction of MRP6 as a transmembrane transporter (Uittoet al., 2001).
The ABCC6 gene comprises 31 exons spanning B73 kb ofgenomic DNA on the short arm of chromosome 16, locus16p13.1. The transcribed messenger ribonucleic acid isB6kb, with a coding sequence of 4.5kb, leading totranslation of a polypeptide with 1503 amino acids. Thefunction of MRP6 is currently unknown; however, it has beenproposed to be a transmembrane transporter, as sequenceanalysis predicts three membrane-spanning domains 1–3 withfive, six, and six transmembrane segments, respectively.
There are two nucleotide-binding folds 1 and 2 (Uitto et al.,2001), and these domains contain conserved Walker A and B motifs critical for the adenosine 5'-triphosphate-binding function, while a conserved C motif, located between the Aand B sequences, is critical for the function of the protein as aputative transmembrane transporter (Borst et al., 1999).
A surprising finding in the context of the widespreadclinical multi-system involvement in PXE was the observationthat MRP6 is expressed predominantly in the liver (Belinskyand Kruh, 1999; Kool et al., 1999). The biological function ofMRP6 in the liver is currently unknown; however, MRP1, theprototype protein within the MRP family, functions as anefflux pump for amphipathic anionic conjugates (Borst et al.,1999). Similarly, MRP6 has been recently shown to transportsmall-molecular-weight glutathione S-conjugates across theplasma membranes in vitro, but the physiological signifi-cance of the substrates being used, including leukotriene C4and ethylmaleimide S-glutathione, is currently unclear(Belinsky et al., 2002; Ilia+s et al., 2002).
All mutations in the ABCC6 gene disclosed in familieswith PXE thus far are recessive, and most of them arepremature termination codon-causing mutations, that is,either nonsense mutations, small insertions or deletionsresulting in frameshift, or large deletions (Le Saux et al.,2001; Ringpfeil et al., 2001a, b; Uitto et al., 2001; Mikschet al., 2005). Also, a number of missense mutations affectingcritical conserved amino acids within the nucleotide-bindingfolds have been disclosed as resulting in functional nullalleles. Thus, most of the cases with classic PXE result fromrecessive null mutations in the ABCC6 gene. However, in anumber of patients with PXE, sequencing of the entire codingregion and splice junctions failed to disclose pathogeneticmutations, raising the possibility that mutations in theregulatory regions of the ABCC6 gene may underlie somecases. Furthermore, it has been suggested that some, but notall, heterozygous carriers of the mutations may show signssuggestive of PXE (Bacchelli et al., 1999; Sherer et al., 2001).
The latter observations raise the issue of pathologicalconsequences of subtle modulation of the ABCC6 geneexpression.
Little is known about the transcriptional regulation of thehuman ABCC6 gene. A recent study (Ara+nyi et al., 2005) hasidentified a methylation-dependent activator sequence in the 50regulatory region of ABCC6, but nothing beyond that hasbeen reported. In the present study, we have provided acomprehensive examination of the transcriptional regulationand tissue-specific expression of the human ABCC6 gene.
RESULTS
High level of expression of the ABCC6 gene in hepatoma cells
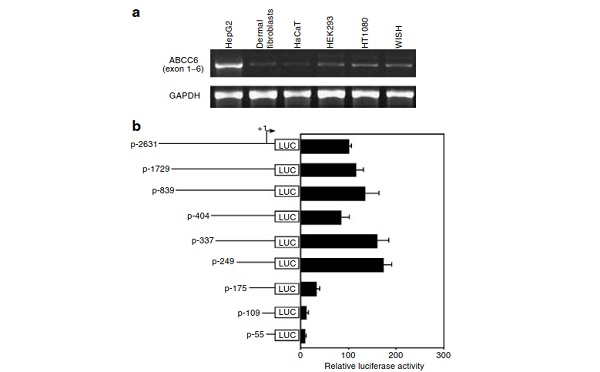
HepG2, a human hepatoma cell line, is expected to expressABCC6, since human liver shows a relatively high level ofexpression of this gene (Belinsky and Kruh, 1999; Kool et al.,1999). We first performed reverse transcriptase-PCR withRNA isolated from HepG2 cells, as well as from a number ofother cultured cells of mesenchymal or epithelial origin,including dermal fibroblasts, HaCaT (a transformed epider-mal cell line), human embryonic kidney (HEK)293 (a kidneyembryonic cell line), HT1080 (a fibrosarcoma cell line), andWISH (an amnionic epithelial cell line) cells. Utilization ofprimers corresponding to exons 1–6 resulted in a strong bandof the expected size (B650 base pairs (bp)) in HepG2 cells,and a much weaker band of the same size was noted in theother cell lines tested (Figure 1a).
Identification of a putative liver-specific cis-element in theABCC6 promoter
To search for regulatory cis-elements that might be critical forexpression of the ABCC6 gene in HepG2 cells, we developeda series of 50deletion constructs consisting of ABCC6promoter linked to luciferase reporter gene. The largestconstruct had its 50end at –2631 (p-2631 construct) upstreamfrom the transcription initiation site ( + 1; Figure 1b). Thetranscription initiation site was determined by 5'-rapid amplification of cDNA ends (5'-RACE), which revealed thatthe transcription site ( + 1) resides 30 nucleotides upstreamfrom the A in the ATG translation initiation codon of thegene. Transfection of the 50deletion constructs into HepG2cells in culture revealed robust expression with the constructp-2631 as well as with constructs containing deletions down to 249 (Figure 1b). However, further deletion of the 50sequences to 175 resulted in about 80% reduction of theluciferase activity, and further reduction was noted withp-109 and p-55 constructs. These observations suggest thatthe segment between 249 and 176 contains elements thatconfer high level of expression of the ABCC6 gene in HepG2cells.
To examine whether the sequence 249/ 176 is aspecific requirement for high level of expression in HepG2cells, we compared the activity of three constructs (p-2631,p-249, and p-175) in HepG2 cells with that in HEK293 andfibrosarcoma cells (HT1080), which expressed ABCC6 at avery low level (see Figure 1a). Transfection experimentsconfirmed that deletion of the 249/ 176 sequence from thepromoter significantly (480%) reduced the activity inHepG2 cells, while no statistically significant differencewas noted in the two other cell lines tested with theconstructs p-249 and p-175 (Figure 2). It should be notedthat the basal promoter activity with the p-2631 constructwas about 50 times higher in HepG2 cells than in HEK293and HT1080 cells, the relative luciferase activity being43.671.5 106vs 0.970.06 106and 1.070.05 106 in HepG2, HEK293, and HT1080 cells, respectively (mean7standard deviation (SD)), after correction of the transfectionefficiency. Thus, the nucleotide segment 249/ 176 of theABCC6 confers high level of expression in the hepatomaHepG2 cells.
Examination of the nucleotide sequence within the 249/176 segment revealed the presence of an NF-kB-likesequence, which differs from the consensus NF-kB-bindingsite by substitution of T by C in the sixth position of the 10-bpconsensus sequence (GGGAMTNYCC; M + A or C; N + anynucleotide; Y + C or T) (see Figure 3). No homology withpreviously published liver-specific cis-elements was notedwithin this segment (Hayashi et al., 1999).
To examine for the presence of transacting factors thatmight bind to the NF-kB-like sequence, an 18-bp probe,239/ 222, was synthesized and used in electrophoreticmobility shift assay (EMSA) with nuclear proteins isolatedfrom HepG2 cells. In addition to nonspecific bands, aradiolabeled DNA/protein complex was noted in electro-phoresis (Figure 3). Addition of unlabeled probe 239/ 222as a competitor abolished this binding in 100-fold excess,while the nonspecific complexes were not affected. How-ever, addition of a 22-bp unlabeled oligonucleotide contain-ing the consensus NF-kB-binding site, in 100-fold excess,failed to significantly reduce the specific DNA/proteincomplex and did not affect the nonspecific binding (Figure 3).
Nevertheless, slight reduction in the intensity of the specificcomplex was noted with 35'-fold excess of the NF-kB oligo-mer (not shown). To examine the sequence requirements for nuclear protein binding within the NF-kB-like sequence infurther detail, three mutant oligomers (M1–M3) were synthe-sized, which harbored 3-bp substitutions affecting differentparts of the wild-type (WT) sequence (Figure 3). Using theseoligomers as competitors in 100-fold excess revealed that M1and M2 probes were able to compete for the protein bindingto the same extent as the WT probe (Figure 3b). However,mutations in probe M3 abolished its ability to compete forprotein binding. Collectively, these findings suggest thatthe NF-kB-like sequence provides high level of expressionin liver cells to the ABCC6 promoter. It should be notedthat two putative CCAAT/enhancer-binding proteincognate sites (Hayashi et al., 1999) were identified atpositions 375 to 366 and 336 to 330. However,elimination of these sequences (compare constructs p-404,p-337, and p-249) did not alter the ABCC6 promoter activity(Figure 1b).
The function of promoter in vivo
To further study the liver-specific characteristics of the humanABCC6 promoter, we examined the activities of the full-length (p-2631) and the shorter (p-249) promoter–luciferaseconstructs in vivo, utilizing a rapid tail vein injectiontechnique (Zhang et al., 1999) (Figure 4). An efficient genetransfer can be achieved in mouse liver by a rapid tail veininjection of a large volume of plasmid DNA solution(hydrodynamics-based transfection). The gene transfer effi-ciency was confirmed by b-galactosidase staining of liverafter transfer of pCMV-LacZ construct into liver with thistechnique (Figure 4b). Injections of 100 mg ABCC6 promoter–luciferase construct DNA in 2 ml Ringer’s solution wereperformed on sets of three mice per construct. The resultsshown in Figure 4a indicate that both promoters arefunctional in vivo, and demonstrate significantly elevatedactivity compared to the controls injected with 2 ml ofRinger’s solution alone. However, the full-length promoterconstruct (p-2631) yielded significantly higher luciferaseexpression and was about 13.6-fold higher than the expres-sion of the p-249 construct. In vitro, the ratio of expression ofthese two constructs was B0.7. This may reflect the fact thatgene regulation in vivo is a more complex process than inisolated cells in culture, and more regulatory elements/factorsare probably involved in the expression of ABCC6 in vivo.
Search for binding proteins on the promoter
For a systematic search of the transcription factors that bind tothe ABCC6 transcriptional regulatory region, we used thep-2631 construct consisting of ABCC6 promoter linked toluciferase reporter gene to isolate a 2661-bp DNA fragmentspanning nucleotide 2631 to + 30. This fragment was usedto isolate nuclear proteins which bind to this region ofpromoter, and the identities of the transcription factors weredetermined by a protein/DNA array-based procedure, asdescribed in Materials and Methods. The results, shown inFigure 5, identified a total of 18 transcription factors in thepulled-down samples. However, among them, only fourfactors were found to have the corresponding consensus-binding sequences within this region of DNA, based oncomputer searches, viz., activator protein-2, USF-1, NF-kB,and epidermal growth receptor (Figure 9).
Cytokine modulation of ABCC6 gene expression and the roleof Sp1/Sp3 in transforming growth factor-b responsiveness
Although there is no evidence that PXE involves tissueinflammation, PXE-like cutaneous findings have been en-countered in a number of metabolic, both acquired andheritable, disorders, some of which involve immunologic andinflammatory aberrations (Ringpfeil and Uitto, 2005). Sinceinflammatory cytokines play a role in a number ofpathological conditions by regulating the expression of genesat the transcriptional level, we examined the prototypic pro-inflammatory cytokines, transforming growth factor (TGF)-b,tumor necrosis factor (TNF)-a, and interferon (IFN-g), for theireffects on the ABCC6 promoter activity in HepG2 cellstransiently transfected with the p-2631 promoter–reportergene construct. The results indicated that addition of TGF-b(0.1–10 ng/ml) increased the promoter activity in a dose-dependent manner up to about 2.5-fold (Figure 6a). At thesame time, addition of TNF-a (0.1–10 ng/ml) or IFN-g(10–1,000 U/ml) suppressed the promoter activity (Figure6a). Further analysis utilizing the 5'-deletion constructsrevealed that the constructs with their 5'-end either at 337or at 175 similarly responded to TGF-b (10 ng/ml),depicting over two-fold increase. Careful search for sequencehomology for known TGF-b response elements, including theSMAD-binding site (Shi et al., 1998; Zawel et al., 1998),revealed the presence of a CAGA box-like sequence,CAGACAGA, superimposed on the transcription initiationsite ( 3 to + 7) (Figure 7). Furthermore, at the position 58to 49 upstream from the transcription initiation site, therewas a consensus Sp1-binding site (Figure 7). To examine the role of these two cis-elements in TGF-b responsiveness,discrete 2- or 4-bp mutations were introduced into thesesequences within the promoter–reporter gene constructp-337, and the WT and mutated promoters were transfectedto HepG2 cells, followed by incubation with or withoutTGF-b (10 ng/ml) (Figure 7). In accordance with the findingsreported above, the WT p-337 promoter construct respondedto TGF-b with about 2.2-fold enhancement of luciferaseactivity. Introduction of 2-bp mutations in the CAGA box-likesequence somewhat reduced, but did not entirely abolish, theTGF-b responsiveness (Figure 7). However, introduction ofmutations to the putative Sp1-binding site entirely abolishedthe upregulation of the promoter activity by TGF-b. Theseobservations suggest that the consensus Sp1-binding siteplays the primary role in TGF-b response within the ABCC6promoter.
To examine the role of the Sp1 cognate sequence infurther detail, EMSA was performed with a radiolabeledprobe extending from 62 to 45 (Figure 8a). Incubation ofHepG2 nuclear extracts with the radiolabeled probe revealeda double band as well as a faster moving band, which wereshown to be specific DNA/protein complexes (Figure 8b, leftarrows). Supershift with antibodies recognizing Sp1 proteinepitopes resulted in disappearance of one of these bands,while two different bands disappeared with the Sp3antibodies with concomitant formation of supershift com-plexes (Figure 8b). The same three bands were dissolved bythe addition of 5'-fold excess of unlabeled 62/ 45oligomer (WT) or addition of an oligomer containing theSp1 consensus-binding site (Figure 8b). Interestingly, additionof the Mut1 62/ 45 probe (M1) containing a 2-bp mutationin Sp1-binding sequence partially competed for the WT62/ 45 probe binding to the nuclear proteins. However,introduction of a 4-bp substitution in the Sp1 sequence(Mut2 62/ 45 (M2) probe) abolished the ability to competefor the WT probe to bind to Sp1/Sp3 proteins. Collectively,these observations attest to the role of Sp1/Sp3 transactingfactors in regulation of ABCC6 gene expression and itsresponsiveness to TGF-β.
Activation of the ABCC6 promoter in Drosophila SL2 cellsby the human Sp1 transcription factor
As Sp1 is expressed in virtually all mammalian cells, weutilized Drosophila SL2 cells, an established in vitro modellacking endogenous SP-1 activity, to determine whether thetranscription factor specifically activates the ABCC6 promo-ter. This was done by transfecting ABCC6 promoterconstructs p-337 and p-249 to SL2 cells. Very low level ofexpression was noted when these reporter gene constructswere expressed together with a control pPAC (-Sp1) vector(Table 1). However, cotransfection of the ABCC6 promoterswith a construct expressing Sp1 full-length cDNA under theDrosophila actin promoter resulted in enhancement ofluciferase activity up to B75-fold (Table 1).
DISCUSSION
The ABCC6 gene encoding MRP6 is expressed primarily inthe liver and to a lesser extent in the kidneys (Belinsky andKruh, 1999; Kool et al., 1999), while very low level ofexpression has been observed in a number of other tissues(Beck et al., 2003). Immunohistochemical staining with antibodies recognizing MRP6 has located it to the basolateralplasma membrane of hepatocytes (Scheffer et al., 2002), afeature consistent with the presence of three membrane-spanning domains in the protein. Highly sensitive multi-round reverse transcriptase-PCR and RNase protection assayapproaches have suggested expression also in other tissues,including skin and vessel wall, albeit at a much lower level(Bergen et al., 2000). Consequently, the gene potentially hastranscriptional cis-elements that confer high liver-specificexpression to this gene. In this study, we first explored liverspecificity of the ABCC6 expression by utilizing HepG2 cells,a hepatoma cell line that has an expression profilecharacteristic of hepatocytes (Khalil et al., 2001). Wedeveloped a number of 50deletion promoter–reporter geneconstructs and transfected them into HepG2 cells, as well asto a number of other established cell lines of mesenchymal orepithelial origin. Transient transfections revealed signifi-cantly, 45'-fold, higher expression of the promoter con-structs in HepG2 cells as compared with HEK293 cells orHT1080, a fibrosarcoma cell line. Thus, the ABCC6 promoterapparently contains features that confer high level ofexpression in the liver.
Initial scanning of the nucleotide sequence informationwithin 2631 bp upstream from the transcription initiation siteof the ABCC6 gene revealed the presence of two sequencemotifs with homology to the consensus CCAAT-binding site.
The corresponding enhancer-binding protein has been shownto be a liver-enriched transcription factor presumablyparticipating in hepatic differentiation and involved both indetermination and maintenance of the hepatic phenotype(Hayashi et al., 1999). Use of 5'-deletion constructs intransient transfection assays revealed, however, that elimina-tion of these CCAAT elements (at 375 to 366 and 336and 330) from the promoter had no appreciable effect onthe promoter activity in HepG2 cells. In contrast, eliminationof the promoter segment between 249 and 176 resulted insignificant (480%) reduction in its activity. This finding wassimilar to that reported by Ara+nyi et al (2005), who notedB50% reduction when the sequence from 332 to 145was eliminated from their promoter construct. The sequencebetween 249 and 176 was found to contain an NF-kB-likesegment ( 235 to 226), which differed from the NF-kBconsensus sequence by one nucleotide substitution. Thissequence was shown to bind, in a specific manner, nuclearproteins isolated from HepG2 cells, and competition assayssuggested the importance of distinct cytosine residues withinthe 30-end of this 10-bp sequence. This NF-kB-like sequencerepresents a novel liver-specific element. It should be notedthat careful sequence analysis failed to identify other,previously established sequences for liver-enriched transcrip-tion factors, including hepatocyte nuclear factors 1, 3, and 4(Hayashi et al., 1999). It is conceivable, therefore, that thepresence of this novel cis-element is responsible for the highlevel of expression of ABCC6 in the liver.
Liver-specific expression of p-2631 construct was alsoalluded to by high level of expression in vivo following itsinjection to the tail vein of normal mice. The rapid injectionof the large volume of fluid enables DNA delivery to the liverby causing a transient right-sided congestive heart failure andbackpressure to the liver vessels (Zhang et al., 1999).
Unexpectedly, the expression of the p-249 construct, whichshowed somewhat higher level of expression in HepG2 cellsin culture, was significantly lower than that of p-2631. Thesefindings may reflect the fact that expression of genes in invivo may be more complex and involves a number ofadditional factors (Figure 9). Nevertheless, our resultsindicate that the expression of the p-2631 construct contain-ing human ABCC6 promoter segment is high both in vitro andin vivo. In this context, it should be noted that human ABCC6has at least two pseudogenes, ABCC6-C1 and ABCC6-C2,that extend from exon 1 to exon 9 and to exon 4, respectively,and contain 5'-flanking sequences. Although ABCC6-C1 is99.995% homologous with ABCC6, there are a number ofdifferences in the 5'-region which allow distinction betweenthe pseudogene and the functional WT gene (Pulkkinen et al.,2001).
We further examined the capacity of the 2631 to + 30promoter region to bind transcription factors using a protein/DNA array. This recently developed technology is asignificant improvement over gel mobility-shift assays, and allows functional analysis of dozens of eukaryotic transcrip-tion factors at a time. This approach identified 18 putativetranscription factors in the sample bound to DNA, but theircounterpart cognate-binding sequences were identified onlyin case of four of them, activator protein-2, USF-1, NF-kB,and epidermal growth receptor. The functionality of NF-kBwas suggested by 50deletion analyses as well as by mutationanalysis altering the NF-kB binding (see Results), while theputative functions of the other three factors are currentlyuntested. It should be noted that computer searches for 14transcription factors identified by the protein/DNA arrayapproach did not find the corresponding cognate-bindingsequences in DNA. This could be explained in some cases bythe possibility that these factors do not directly bind to DNAbut form complexes with other factors. The second explana-tion may reside in the fact that the stringency of the searchdid not allow recognition of binding sites with o80%homology with the consensus sequence. Finally, there maybe some crosshybridization between the families of transcrip-tion factors. For example, while binding signals for PAX-5and hepatocyte nuclear factor-4 were noted (see Figure 5),no consensus-binding sites for these factors were identified.
However, consensus sites for PAX-4 and hepatocytenuclear factor-3b were identified with partial sequencehomologies.
The complexity of the transcriptional regulation of ABCC6has also been attested by a recent study identifying a DNAmethylation-dependent activator sequence in ABCC6 (Ara+nyiet al., 2005). Specifically, these authors identified bothactivator and repressor sequences in the proximal promoterregion. The most potent activator sequence consisted ofconserved elements protected by DNA methylation innonexpressing cells (Ara+nyi et al., 2005). These findings,together with our results, raise the possibility that mutationsin the regulatory regions of the ABCC6 gene may underliesome cases of PXE. However, besides large genomicdeletions affecting the promoter region, no regulatorymutations have been disclosed in the ABCC6 gene (Mikschet al., 2005; our unpublished results).
Another novel observation derived from the present studyis that cytokines, including TGF-b, TNF-a, and IFN-g, are ableto modulate the ABCC6 promoter activity. In particular, TGF-b,a profibrotic cytokine in the liver (Bissell et al., 2001),significantly upregulated the ABCC6 promoter activity.
Scanning of the promoter sequence for putative TGF-bresponse elements identified a CAGA box-like sequence(GACAGACAGA) overlapping the transcription initiation site( 3 to + 7). This segment has similarity to the consensusmotifs for Smad binding (GTCTAGAC, so-called Smad-binding element, and AG(C/A)CAGACAC, so-called CAGAbox). Both sequences contain the core motive AGAC, whichrepresents the optimal binding sequence for Smad3 andSmad4 (Shi et al., 1998; Zawel et al., 1998), a sequence alsopresent in the CAGA box-like sequence in the ABCC6promoter. However, mutation of the CAGA box-likesequence by 2-bp substitutions (see Figure 7) failed to abolishthe TGF-b responsiveness of the promoter. The inability of theCAGA box-like sequence to serve as a functional TGF-βresponse element through Smad binding may relate to itsposition within the gene.
In contrast to the CAGA box-like sequence, an upstreamSp1-binding site at –58 to –49 was shown to be critical forTGF-b response within the ABCC6 promoter. Specifically, 2-or 4-bp substitutions within the Sp1 consensus sequenceentirely abolished the responsiveness to TGF-b. EMSA withsupershift using specific antibodies identified Sp1 and Sp3 asnuclear proteins specifically binding to the Sp1 cognatesequence in the ABCC6 gene promoter. Previous studies havesuggested that Sp1 binding is necessary for TGF-b1-inducedexpression by a number of genes, as exemplified by proa2(I)collagen and b5 integrin (Lai et al., 2000; Poncelet andSchnaper, 2001), and it has been suggested that geneactivation in these cases may involve cooperation betweenSmad3 and Sp1. However, Sp1-binding site has been shownto function as a distinct TGF-b responsive element forpromoter expression and Sp1 by itself can mediate thisresponse (Li et al., 1998). In case of ABCC6 promoter, site-directed mutagenesis of the Sp1 site alone was able toabrogate the TGF-b responsiveness, suggesting a critical rolefor this cis-element in regulation of the correspondinggene expression. The expression of the ABCC6 promoterwas also shown to be dependent on Sp1 protein by theuse of Drosophila SL2 cells. This is an established in vitromodel to study the role of Sp1, since these cells are devoidof endogenous Sp1, while all mammalian cells containthis protein (Courey and Tjian, 1988). Specifically, transfec-tion of two promoter–luciferase constructs (p-337 and p-249),both of which contain the Sp1 sequence at 58 to 49, toSL2 cells resulted in low level of expression. However,cotransfection with a construct expressing Sp1 under theDrosophila actin gene promoter resulted in up to 75-foldenhancement of the ABCC6 promoter activity. Theseobservations clearly attest to the importance of Sp1 inregulation of the ABCC6 promoter activity and its respon-siveness to TGF-b.
MATERIALS AND METHODS
Cell culture
All mammalian cell lines and human dermal fibroblasts, obtainedfrom neonatal foreskin, were cultured in medium supplemented with10% heat-inactivated fetal calf serum, 2 mM L-glutamine, 100 IU/mlpenicillin, and 100 mg/ml streptomycin (Cellgro, Mediatech, Inc.,Herndon, VA). In case of human HepG2 hepatoma cells, themedium was minimal essential medium (Cellgro), while humandermal fibroblasts, HaCaT-transformed epidermal cells, HEK293cells, HT1080 fibrosarcoma cells, and WISH amniotic cells werecultured in Dulbecco’s modified Eagle’s medium (Cellgro). Cultureswere maintained at 371C in a humidified atmosphere of 5% CO2and95% air.
Drosophila SL2 cells (kindly provided by Dr James Jaynes,Thomas Jefferson University, Philadelphia, PA) were grown at roomtemperature in Drosophila Schneider cell medium (Gibco, Carlsbad,CA) supplemented with 10% heat-inactivated fetal bovine serum.
The experiments were approved by the Institutional ReviewBoard, at Thomas Jefferson University, and they adhere to theDeclaration of Helsinki Principles.
Reverse transcription-PCR
Total RNA was isolated from cultured cells using TRIzol reagent, asrecommended by the manufacturer (Invitrogen, Carlsbad, CA). First-strand cDNA synthesis was performed using 1 mg total RNA, oligo(dT) primer (Promega, Madison, WI), and Superscript II reversetranscriptase (Invitrogen) according to the manufacturer’s instruc-tions. The following primers were used: 5'-ATGGCCGCGCCTGCTGAGCCCTGC-30(sense) and 5'-CCAGTCTCTGGACAGGGGTTAGACTGC-30(antisense) for ABCC6; 5'-GGTGAAGGTCGGAGTCAACGGA-30(sense) and 5'-AGGTCCACCACCCTGTTGCTGT-30(antisense) for glyceraldehyde-3-phosphate dehydrogenase as aninternal control. A 50 ml PCR reaction mixture consisted of 1 PCRbuffer, 1 Q-buffer, 2.5 U Taq polymerase (Qiagen, Valencia, CA),200 mM nucleotide mix, 15 pmol each primer, and 1 ml of reactionmixture containing first-strand cDNA. The amplification conditionswere 941C for 5 minutes, followed by 38 cycles of 941C for45 seconds, 681C for 45 seconds, and 721C for 1 minute, and onecycle of 721C for 10 minutes. PCR products were separated by gelelectrophoresis on 1.5% agarose gels and stained with ethidiumbromide.
5'-rapid amplification of cDNA ends analysis
5'-rapid amplification of cDNA ends (RACE) analysis was performedwith SMART RACE cDNA amplification kit (Clontech Laboratories,Inc., Palo Alto, CA) using 1 mg total RNA isolated from the humanHepG2 hepatoma cells. First-strand cDNA synthesis was performedusing Superscript II reverse transcriptase (Invitrogen), SMART II oligo-nucleotide, and 5'-RACE cDNA synthesis primer, according to themanufacturer’s instructions (Clontech Laboratories, Inc.). 5'-RACEPCR was performed using a universal primer contained in the kit,ABCC6 gene-specific primer (MRP6-1:5'-CCAGTCTCTGGACAGGGGTTAGACTGC-30) located in exon 5, and Advantage 2 polymer-ase. The amplified products were diluted 5'-fold, and a volume of1 ml was used in nested 5'-RACE with nested universal primer and thesecond ABCC6 gene-specific primer (MRP6-2: 5'-GGAACACTGCGAAGCTCATCGTGG-30) located at the exon 3–4 border, according tothe manufacturer’s instructions (Clontech Laboratories, Inc.). Theproducts were cloned into the pCR4-TOPO vector as recommendedby the manufacturer (Invitrogen). Recombinant plasmids werepurified with Miniprep Kit (Qiagen) and subjected to nucleotidesequence analysis using ABI 377 DNA sequencer.
Promoter plasmid constructs
A 2661-bp fragment spanning from 2631 to + 30 of the promoterregion of the human ABCC6 gene was prepared by PCR amplifica-tion of the human genomic DNA using a sense primer ( 2631F: 5'-GTGGTACCAAGGCGTACAGCCACTGTGA-30) containing a KpnIrestriction site and an antisense primer ( + 30R: 5'-TACTCGAGTTCTGTCGTCGTGGGTCCCAGCGT-30 ) containing an XhoI restrictionsite. The PCR products were separated by agarose gel electrophoresisand extracted from a gel slice (Qiagen). The purified fragment wasdigested with KpnI and XhoI, and cloned into pGL3 basic luciferasevector (Promega) between KpnI and XhoI sites to generate the p-2631 construct. Additional reporter gene constructs containingsequentially truncated fragments from the 5'-end of p-2631, span-ning from 1729, 839, 404, 337, 249, 175, 109, and 55to + 30 of the ABCC6 promoter region were similarly prepared usingsense primers containing a KpnI restriction site and the antisense primer + 30R. Mutagenesis of the Sp1-binding site and CAGA box-like sequence in p-337 reporter construct, spanning from 337 to+ 30, was performed by site-directed mutagenesis, as described byHo et al. (1989) using a sense primer ( 337F: 5'-GCGGTACCTGGAAATTGCTGGGTCCA-30), antisense primers ( ? 30R-1mCAGA: 5'-TACTCGAGTTCTGTCGTCGTGGGTCCCAGCGTCAATCTG-30, ? 30R-2mCAGA: 5'-TACTCGAGTTCTGTCGTCGTGGGTCCCAGCGTCTGAATG-30) containing an XhoI restriction site, and the mutant oligo-nucleotides (mutated nucleotides are underlined in bold). The finalconstructs were sequenced in both directions to ensure correctnucleotide sequence. The sequence of the insert in p-2631 constructconfirmed its fidelity with human ABCC6 database sequence(Figure 9). Computer analysis of the promoter region of ABCC6was conducted to detect putative cis-acting elements usingtranscription factor databases (TFSEARCH, Kyoto University, version1.3) and ConSite (nsiteM, Softberry, Inc., version 2.2004; www.phy-lofoot.org).
Transient transfections and luciferase assay
Plasmid constructs used for transient transfections were preparedusing a purification kit (Qiagen). HepG2, HEK293, and HT1080 cellswere plated on 35-mm dishes 24 hours prior to transfection andgrown to approximately 80% confluency. The cells were transfectedwith 0.8 mg of experimental plasmid and 0.2 mg of pRSV-b-galactosidase plasmid as an internal control of transfectionefficiency, using FuGENE 6 transfection reagent according to themanufacturer’s instructions (Roche Diagnostic Co., Indianapolis, IN).
For Sp1 cotransfections in Schneider Drosophila SL2 cells, the sametransfection methods were used, except that 0.4 mg of the ABCC6promoter–luciferase constructs were cotransfected with 0.4 mg ofpPACSp1 (Sp1 expression vector) or pPAC (empty vector as control),which were a generous gift from Dr Robert Tjian, University ofCalifornia, Berkeley. In addition, in each experiment, 0.2 mg ofpHSPLacZ was used as an internal control of transfection efficiency(Kadonaga et al., 1987).
For experiments with cytokines, HepG2 cells were washed twicewith sterile phosphate-buffered saline 18 hours after transfection andthen incubated in serum-free minimal essential medium for 6 hoursprior to addition of the cytokines for 24 hours. Incubations wereperformed with human TGF-b (R&D systems, Minneapolis, MN),human TNF-a (R&D systems), and human interferon-g (IFN-g, RocheDiagnostic Co.).
The transfected cells were harvested in reporter lysis buffer(Promega) and used to measure the luciferase activity with theLuciferase Assay Reagent (Promega) using Lumat LB 9507 lumin-ometer (Berthold, Wildbad, Germany). The b-galactosidase activitywas determined according to standard protocols (Sambrook et al.,1989), and luciferase activity (arbitrary units) was pided byb-galactosidase activity in the same sample (densitometric units at420 nm) to correct for transfection efficiency and expressed asrelative luciferase activity. Luciferase assays were carried out intriplicate, and each experiment of transfection was repeated at leastthree times. Data shown in the figures represent the mean7SD ofthree independent experiments.
To investigate the activity of ABCC6 promoter–luciferase reportergene construct in vivo, B3.5-month-old male FVB/N mice wereused for tail vein injections with the constructs. A CMV–lacZconstruct was injected as a positive control to check gene transferefficiency to different tissues, and Ringer’s solution was injected as anegative control. The injections were peformed using a 27-gaugeneedle and 100 mg of each DNA construct in 2 ml of Ringer’ssolution within 7 seconds. Several tissues, including the entire liver,were collected from each injected mouse at day 2 postinjection forluciferase assay or for b-galactosidase staining according to standardprotocols (Manthorpe et al., 1993; Zhang et al., 1999). The luciferaseactivity (arbitrary units) was normalized to per mg of liver extractprotein and expressed as relative luciferase activity. Each assay wasperformed in triplicate.
EMSA
Nuclear extracts were prepared from HepG2 cells according to anestablished protocol (Andrews and Faller, 1991) and stored at 801Cuntil use. The protein concentration in the extracts was determinedby a commercial assay kit (Bio-Rad Laboratories, Hercules, CA).
Double-stranded oligonucleotides ( 239/ 222 and 62/ 45) wereend-labeled with [g-32P]dATP by T4 polynucleotide kinase (Prome-ga). In all, 6 mg of the nuclear extract was incubated in the bindingbuffer (10 mM Hepes (pH 7.6), 4% glycerol, 1% Ficoll, 25 mM KCl,1 mM dithiothreitol, 0.5 mM EDTA, and 25 mM NaCl) containing 1 mgpoly(dI-dC) (Roche Diagnostic Co.) and 20 mg bovine serum albuminon ice for 10 minutes. End-labeled oligonucleotides, 60,000 c.p.m.,were added to the mixture and incubated at room temperature for20 minutes. For competition experiments, 5'- or 100-fold molarexcess of unlabeled oligonucleotides were added to the bindingreaction mixture 10 minutes prior to the addition of the labeledprobe. Oligonucleotides containing Sp1 or NF-kB consensus-binding site were purchased from Promega. For supershift experi-ments, 2 mg of antibodies against Sp1 or Sp3 (Santa CruzBiotechnology, Santa Cruz, CA) were incubated with the bindingreaction mixture on ice for 1 hour before the labeled probe wasadded. The DNA–protein complexes were separated by electrophor-esis on 4% polyacrylamide gel in 0.5 TBE at 200 V for 2 hours at41C, fixed for 30 minutes in 30% methanol and 10% acetic acid,vacuum-dried, and autoradiographed.
Protein/DNA array
A 2661-bp fragment of ABCC6 promoter region, extending from2631 to + 30, was excised from p-2631 construct with restrictionenzymes KpnI and XhoI, and labeled with biotin using Bio-16-dUTP(Roche, Mannheim, Germany) and the Klenow fragment of DNApolymerase I (Invitrogen). Unincorporated Bio-16-dUTP was re-moved by a spin column and the biotin-labeled DNA fragment wascoupled to the M-280 streptavidin magnetic beads (Dynal Biotech,Oslo, Norway) under the conditions suggested by the manufacturer.
The DNA-coupled magnetic beads were incubated with 500 mgprotein in HepG2 nuclear extracts for 2 hours at 41C in the bindingbuffer (4% Ficoll, 20 mM Hepes, pH 7.9, 1 mM EDTA, 1 mMdithiothreitol, 50 mM KCl, 0.05% Triton X-100, 10% glycerol) withan excess amount of dC-dI for competition of nonspecific binding.
After several washes, the bound proteins were dissociated from theDNA-coupled beads by incubation in a buffer containing 2 M NaCl,for 60 minutes on ice.
The bound proteins extracted from the 2661-bp promoter regionfragment were incubated with the TranSignal (Panomics, Redwood,CA) probe mix, a set of 54 biotin-labeled DNA-binding oligonucleo-tides corresponding to the consensus sequences for the correspond-ing transcription factors, respectively, to allow the formation ofDNA/protein complexes. The transcription factor-bound probeswere isolated and then dissociated from the DNA/protein com-plexes, and used to hybridize to the TranSignal Array that had beenspotted with complementary consensus-binding sequences of thetranscription factor probes, at 421C for 8 hours. Hybridizationsignals were visible after exposure to X-ray film following chemi-luminescent detection. Array hybridization was repeated using thenuclear extracts prepared separately and the same results wereobtained.
CONFLICT OF INTEREST
The authors state no conflict of interest.
ACKNOWLEDGMENTS
We thank Carol Kelly for assistance in the preparation of this manuscript.
Dr James Jaynes (Thomas Jefferson University) and Dr Robert Tjian (Universityof California, Berkley) kindly provided Drosophila cells and Sp1 constructs.
We also thank Dr Lan Huang for technical assistance. These studies weresupported by the USPHS/NIH grant R01AR28450.
REFERENCES
Andrews NC, Faller DV (1991) A rapid micropreparation technique forextraction of DNA-binding proteins from limiting numbers of mamma-lian cells. Nucleic Acids Res 19:2499
Ara+nyi T, Ratajewski M, Bardoczy V, Pulaski L, Bors A, Tordai A et al. (2005)Identification of a DNA methylation-dependent activator sequence in thepseudoxanthoma elasticum gene, ABCC6. J Biol Chem 280:18643–50
Bacchelli B, Quaglino D, Gheduzzi D, Taparelli F, Boraldi F, Trolli B et al.(1999) Identification of heterozygote carriers in families with a recessiveform of pseudoxanthoma elasticum (PXE). Mod Pathol 12:1112–23
Beck K, Hayashi K, Nishiguchi B, Le Saux O, Hayashi M, Boyd CD (2003) Thedistribution of Abcc6 in normal mouse tissues suggests multiplefunctions for this ABC transporter. J Histochem Cytochem 51:887–902
Belinsky MG, Chen Z-S, Shchaveleva I, Zeng H, Kruh GD (2002) Characterization of the drug resistance and transport properties ofmultidrug resistance protein 6 (MRP6, ABCC6). Cancer Res 62:6172–7
Belinsky MG, Kruh GD (1999) MOAT-E (ARA) is a full-length MRP/cMOATsubfamily transporter expressed in kidney and liver. Br J Cancer80:1342–9
Bergen AA, Plomp AS, Schuurman EJ, Terry S, Breuning M, Dauwerse H et al.(2000) Mutations in ABCC6 cause pseudoxanthoma elasticum. NatGenet 25:228–31
Bissell DM, Roulot D, George J (2001) Transforming growth factor beta andthe liver. Hepatology 34:859–67
Borst P, Evers R, Kool M, Wijnholds J (1999) The multidrug resistance proteinfamily. Biochim Biophys Acta 1461:347–57
Courey A, Tjian R (1988) Analysis of Sp1 in vivo reveals multiple transcriptiondomains, including a novel glutamine-rich activation motif. Cell55:887–8
Hayashi Y, Wang W, Ninomiya T, Nagano H, Ohta K, Itoh H (1999) Liverenriched transcription factors and differentiation of hepatocellularcarcinoma. Mol Pathol 52:19–24
Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directedmutagenesis by overlap extension using the polymerase chain reaction.Gene 77:51–9
Ilia+s A, Urba+n Z, Seidl TL, Le Saux O, Sinko E, Boyd CD et al. (2002) Lossof ATP-dependent transport activity in pseudoxanthoma elasticum-associated mutants of human ABCC6 (MRP6). J Biol Chem 277:16860–7
Kadonaga J, Carner C, Masiarz F, Tjian R (1987) Isolation of a cDNA encodingtranscription factor Sp1 and functional analysis of the DNA bindingdomain. Cell 51:1079–90
Khalil M, Shariat-Panahi A, Tootle R, Ryder T, McCloskey P, Roberts E et al.(2001) Human hepatocyte cell lines proliferating as cohesive spheroidcolonies in alginate markedly upregulate both synthetic and detoxifica-tory liver function. J Hepatol 297:68–77
Kool M, van der Linden M, de Haas M, Baas F, Borst P (1999) Expression ofhuman MRP6, a homologue of the multidrug resistance protein geneMRP1, in tissues and cancer cells. Cancer Res 59:175–82
Lai C-F, Feng X, Nishimura R, Teitelbaum SL, Avioli LV, Ross FP et al. (2000)Transforming growth factor-beta up-regulates the beta 5 integrin subunitexpression via Sp1 and Smad signaling. J Biol Chem 275:36400–6
Le Saux O, Beck K, Sachsinger C, Silvestri C, Treiber C, Goring HH et al.(2001) A spectrum of ABCC6 mutations is responsible for pseudox-anthoma elasticum. Am J Hum Genet 69:749–64
Li J-M, Datto MB, Shen X, Hu PP, Yu Y, Wang X-F (1998) Sp1, but not Sp3,functions to mediate promoter activation by TGF-beta through canonicalSp1 binding sites. Nucleic Acids Res 26:2449–56
Manthorpe M, Cornefert-Jensen F, Hartikka J, Felgner J, Rundell A, MargalithM et al. (1993) Gene therapy by intramuscular injection of plasmidDNA: studies on firefly luciferase gene expression in mice. Human GeneTher 4:419–31
Miksch S, Lumsden A, Guenther UP, Foernzler D, Christen-Zach S, DaughertyC et al. (2005) Molecular genetics of pseudoxanthoma elasticum: typeand frequency of mutations in ABCC6. Hum Mutat 26:235–48
Poncelet A-C, Schnaper HW (2001) Sp1 and Smad proteins cooperate tomediate transforming growth factor-beta 1-induced alpha 1(I) collagenexpression in human glomerular mesangial cells. J Biol Chem276:6983–92
Pulkkinen L, Nakano A, Ringpfeil F, Uitto J (2001) Identification of ABCC6pseudogenes on human chromosome 16p: implications for mutationdetection in pseudoxanthoma elasticum. Hum Genet 109:356–65
Ringpfeil F, Nakano A, Uitto J, Pulkkinen L (2001b) Compound heterozygosityfor a recurrent 16.5-kb Alu-mediated deletion mutation and single-base-pair substitutions in the ABCC6 gene results in pseudoxanthomaelasticum. Am J Hum Genet 68:642–52
Ringpfeil F, Pulkkinen L, Uitto J (2001a) Molecular genetics of pseudox-anthoma elasticum. Exp Dermatol 10:221–8
Ringpfeil F, Uitto J (2005) Heritable disorders of connective tissue. In:Dermatology, 2nd edn (Bologna et al. eds), London, UK: Elsevier(in press)
Sambrook J, Fritsh EF, Maniatis T (1989) Molecular Cloning, A LaboratoryManual. 2nd edn. Cold Spring Harbor, NY: Cold Spring HarborLaboratory, 1166–7
Scheffer GL, Hu X, Pijnenborg AC, Wijnholds J, Bergen AA, Scheper RJ (2002)MRP6 (ABCC6) detection in normal human tissues and tumors. LabInvest 82:515–8
Sherer DW, Bercovitch L, Lebwohl M (2001) Pseudoxanthoma elasticum: significance of limited phenotypic expression in parents of affectedoffspring. J Am Acad Dermatol 44:534–7
Shi Y, Wang Y-F, Jayaraman L, Yang H, Massague+ J, Pavletich NP (1998)Crystal structure of a Smad MH1 domain bound to DNA: insight on DNAbinding in TGF-beta signaling. Cell 94:585–94
Uitto J, Pulkkinen L, Ringpfeil F (2001) Molecular genetics of pseudoxantho-ma elasticum—a metabolic disorder at the environment–genome inter-face+ Trends Mol Med 7:13–7
Zawel L, Le Dai J, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B et al.(1998) Human Smad3 and Smad 4 are sequence-specific transcriptionactivators. Mol Cell 1:611–7
Zhang G, Budker V, Wolff JA (1999) High levels of foreign gene expression inhepatocytes after tail vein injections of naked plasmid DNA. Hum GeneTher 10:1735–7